rm(list=ls())
library(pheatmap)
library(Rtsne)
library(mvtnorm)
# 生成数据(随机)================================
# 设置基因数和细胞数
gene_num=100
cell_num=50
# 设置两个正态分布的随机矩阵(100*50) 作为两个样本矩阵
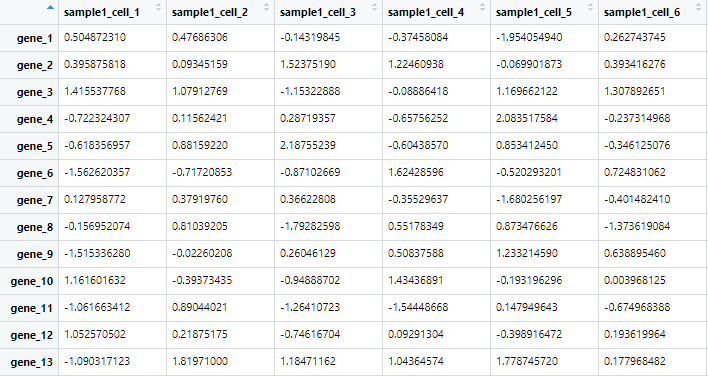
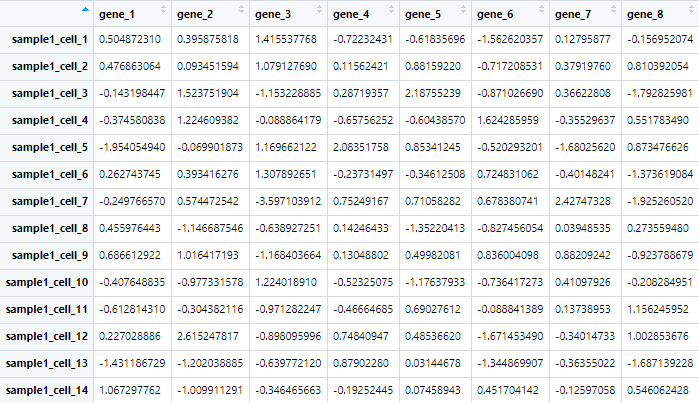
sample1=rnorm(gene_num*cell_num);
# 生成 100 基因,50 细胞的表达矩阵
dim(sample1)=c(gene_num,cell_num)
# 生成第二个样本
sample2=rnorm(gene_num*cell_num);dim(sample2)=c(gene_num,cell_num)
# 将两个样本合并为一个表达矩阵
sample_all=cbind(sample1,sample2)
# 添加列名
colnames(sample_all)=c(paste0('sample1_cell_',1:cell_num),
paste0('sample2_cell_',1:cell_num))
# 添加行名
rownames(sample_all)=paste('gene_',1:gene_num,sep = '')
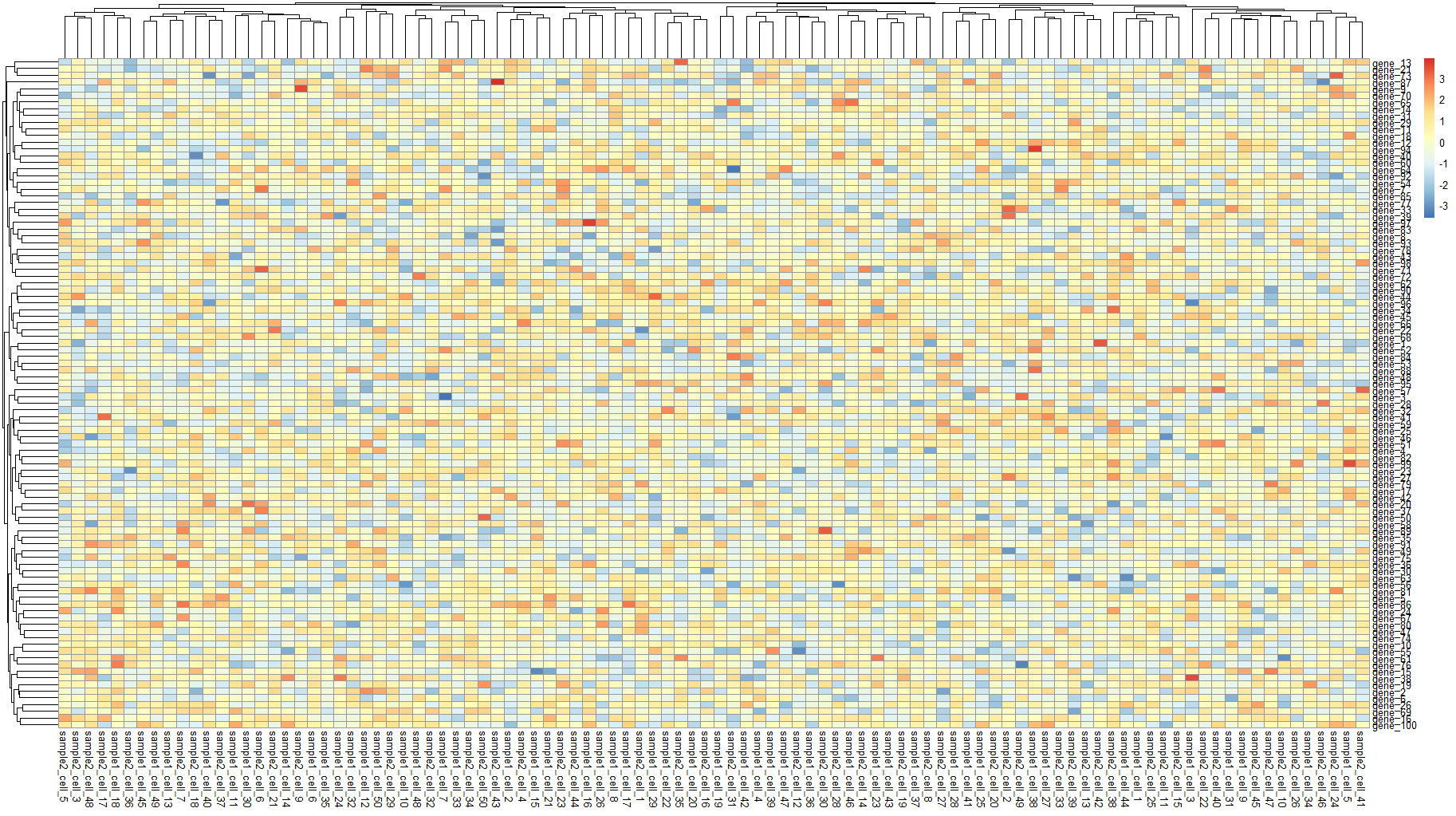
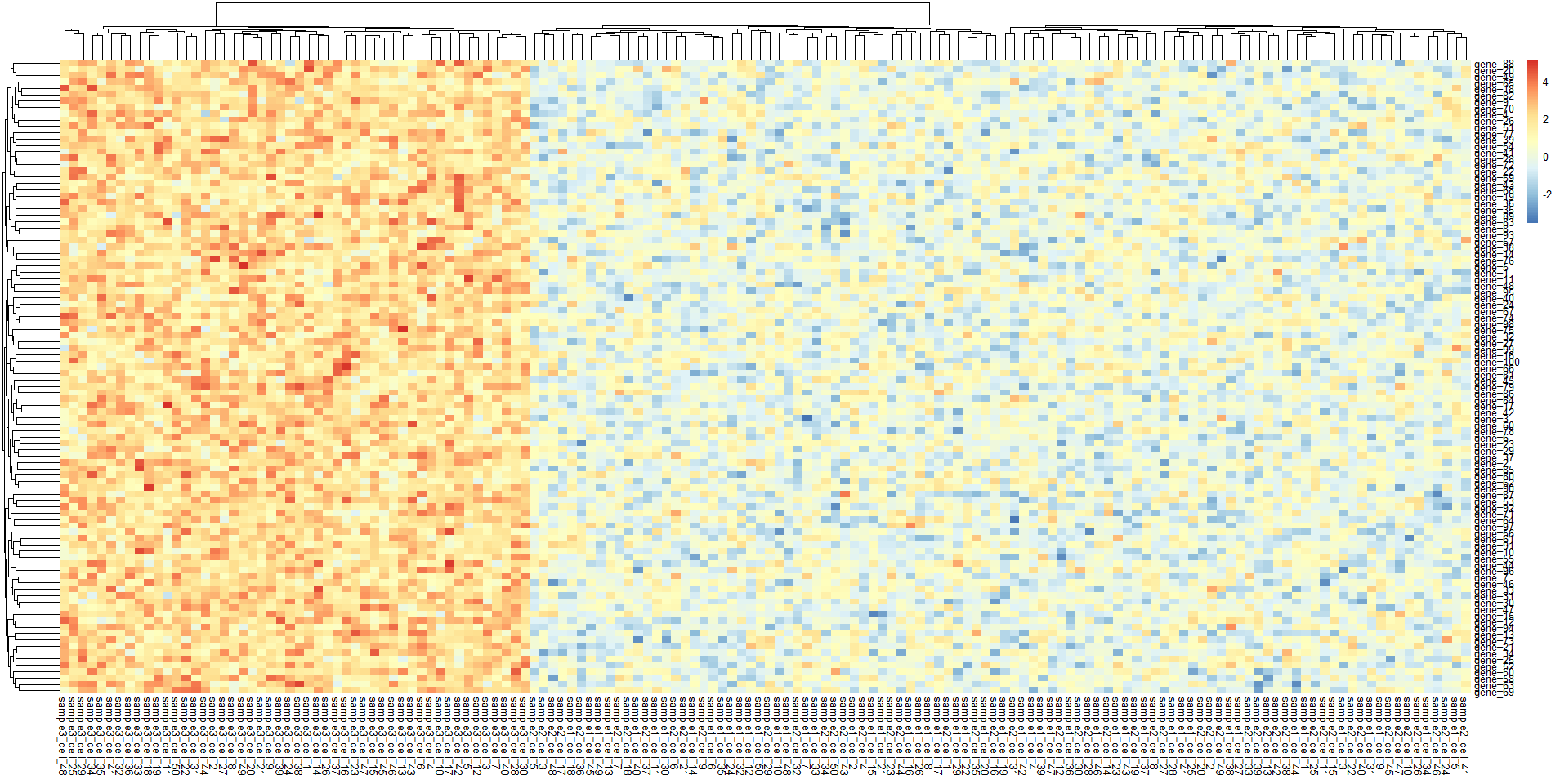
# 绘制热图==============================================
pheatmap(sample_all)
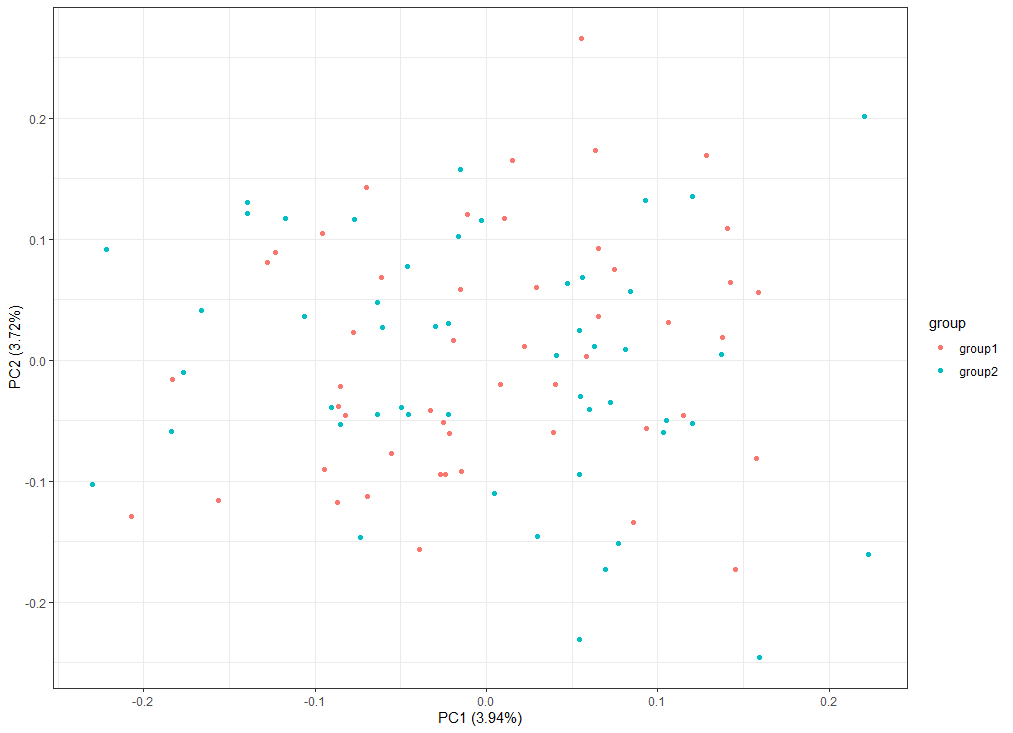
# 绘制pca===============================================
# 转置,让行为样本
sample_all=t(sample_all)
# 添加样本分组信息
df=cbind(as.data.frame(sample_all),group=c(rep('group1',cell_num),rep('group2',cell_num)))
# prcomp()主成分分析
pca_dat <- prcomp( df[,1:(ncol(df)-1)] )
autoplot(pca_dat, data=df,colour = 'group')+theme_bw()
# 绘制tsne=============================================
# 进行tSNE
set.seed(123123)
sample_all=t(sample_all)
tsne_out <- Rtsne(sample_all,pca=FALSE,perplexity=10,theta=0.0)
# 获取tSNE的坐标值
str(tsne_out)
# 其中在Y中存储了绘制图坐标
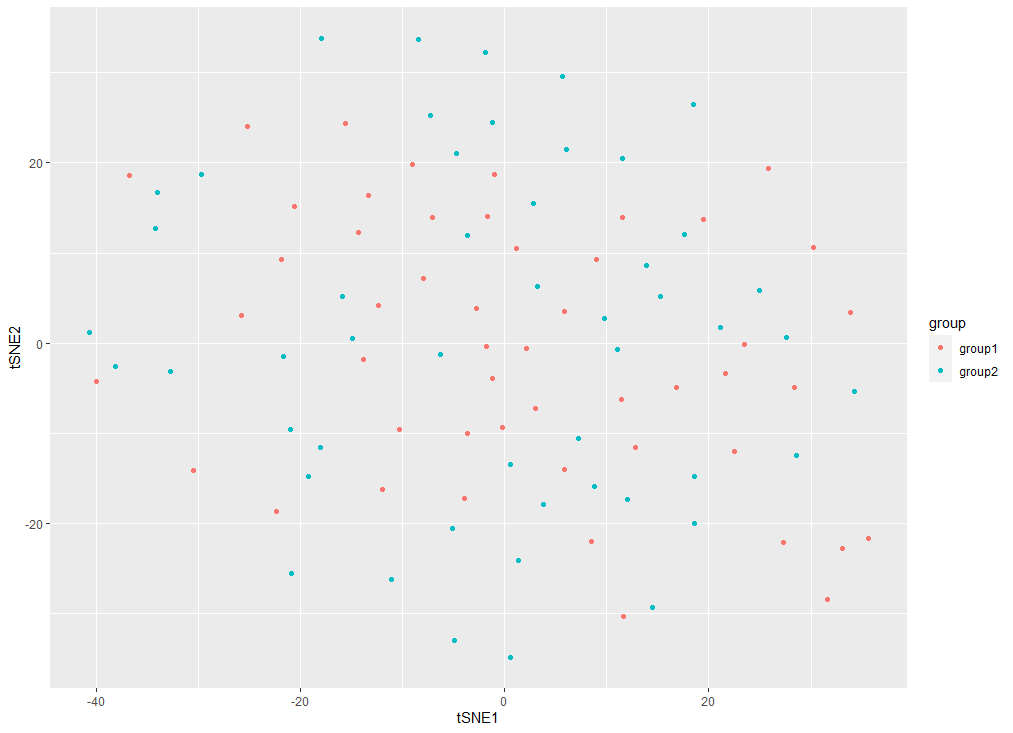
tsnes=tsne_out$Y
colnames(tsnes) <- c("tSNE1", "tSNE2") #为坐标添加列名
# 在此基础上添加颜色分组信息,首先还是将tsnes这个矩阵变成数据框,然后增加一列group信息,最后映射在geom_point中
tsnes=as.data.frame(tsnes)
group=c(rep('group1',cell_num),rep('group2',cell_num))
tsnes$group=group
ggplot(tsnes, aes(x = tSNE1, y = tSNE2))+ geom_point(aes(col=group))
dev.off()
dev.new()
# 构建随机数据集==================================================
# 第三个样本中表达量每个值加2
sample3=rnorm(gene_num*cell_num)+2;dim(sample3)=c(gene_num,cell_num)
colnames(sample3)=c(paste0('sample3_cell_',1:cell_num))
rownames(sample3)=paste('gene_',1:gene_num,sep = '')
sample_all=cbind(sample_all,sample3)
# 绘制热图 =========================================================
pheatmap(sample_all)
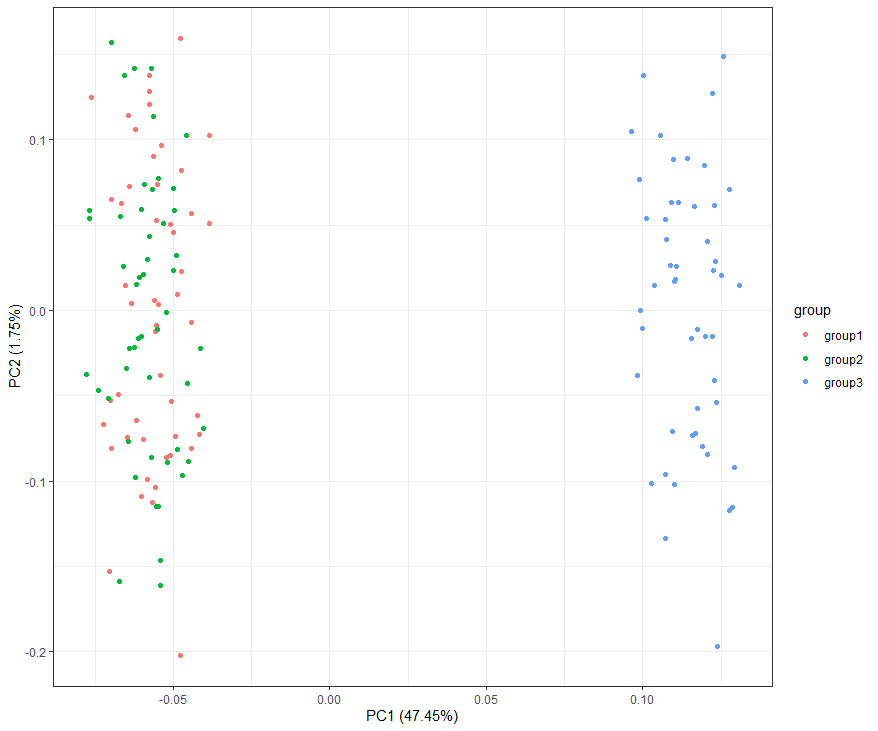
# 绘制PCA ==========================================================
sample_all=t(sample_all);
dim(sample_all)
df=cbind(as.data.frame(sample_all),group=c(rep('group1',cell_num),rep('group2',cell_num),rep('group3',cell_num)))
autoplot(prcomp( df[,1:(ncol(df)-1)] ), data=df,colour = 'group')+theme_bw()
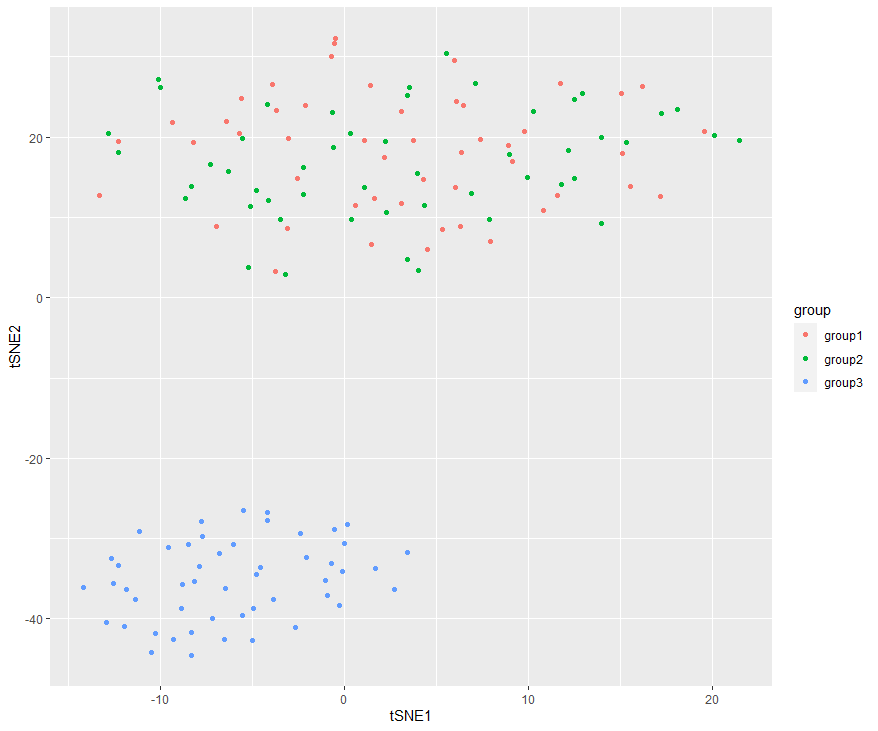
# 绘制tSNE=========================================================
set.seed(123123)
tsne_out <- Rtsne(sample_all,pca=FALSE,perplexity=10,theta=0.0)
tsnes=tsne_out$Y
colnames(tsnes) <- c("tSNE1", "tSNE2")
tsnes=as.data.frame(tsnes)
group=c(rep('group1',cell_num),rep('group2',cell_num),rep('group3',cell_num))
tsnes$group=group
ggplot(tsnes, aes(x = tSNE1, y = tSNE2))+ geom_point(aes(col=group))
|